Sitemap
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Pages
Posts
portfolios
Causality-Induced Positional Encoding for Transformer-Based Representation Learning of Non-Sequential Features
CAPE is a novel positional encoding that identifies underlying causal structure over non-sequential features to improve the performance of transformer-based models. 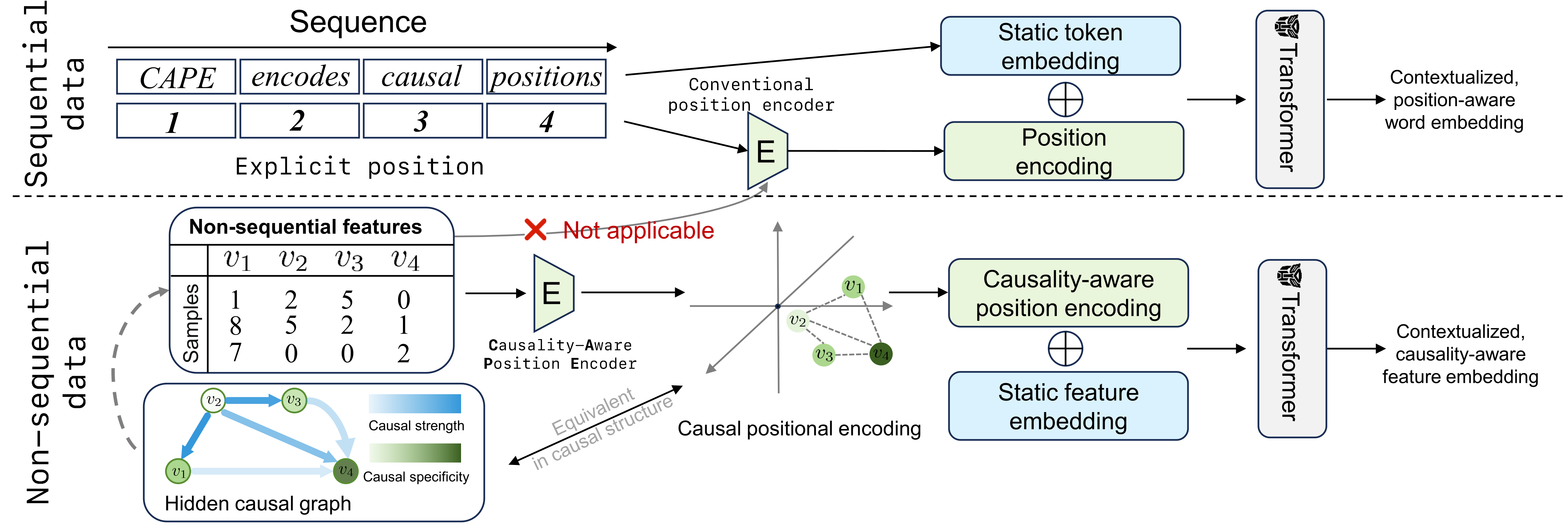
Multimodal Anomalous Tissue Region Detection Enhanced with Spatial Transcriptomics
MEATRD is a multimodal anomaly detection method that integrates histology image and Spatial Transcriptomics gene expression data. 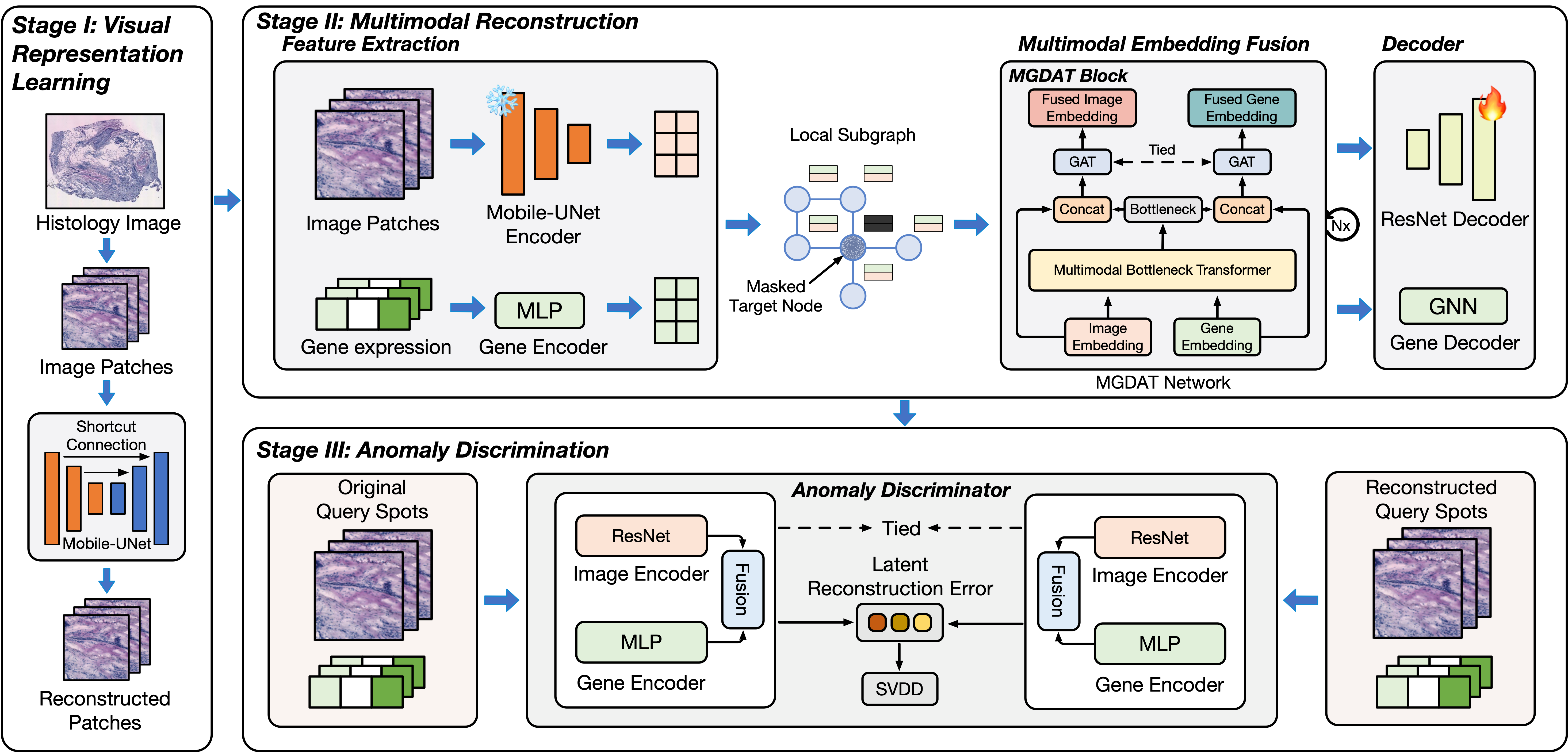
Detecting anomalous anatomic regions in spatial transcriptomics with STANDS
STANDS is a GAN-based multi-task deep learning framework, which can detect and dissect anomalous tissue domains (DDATD) with Spatial Transcriptomics or scRNA-seq. 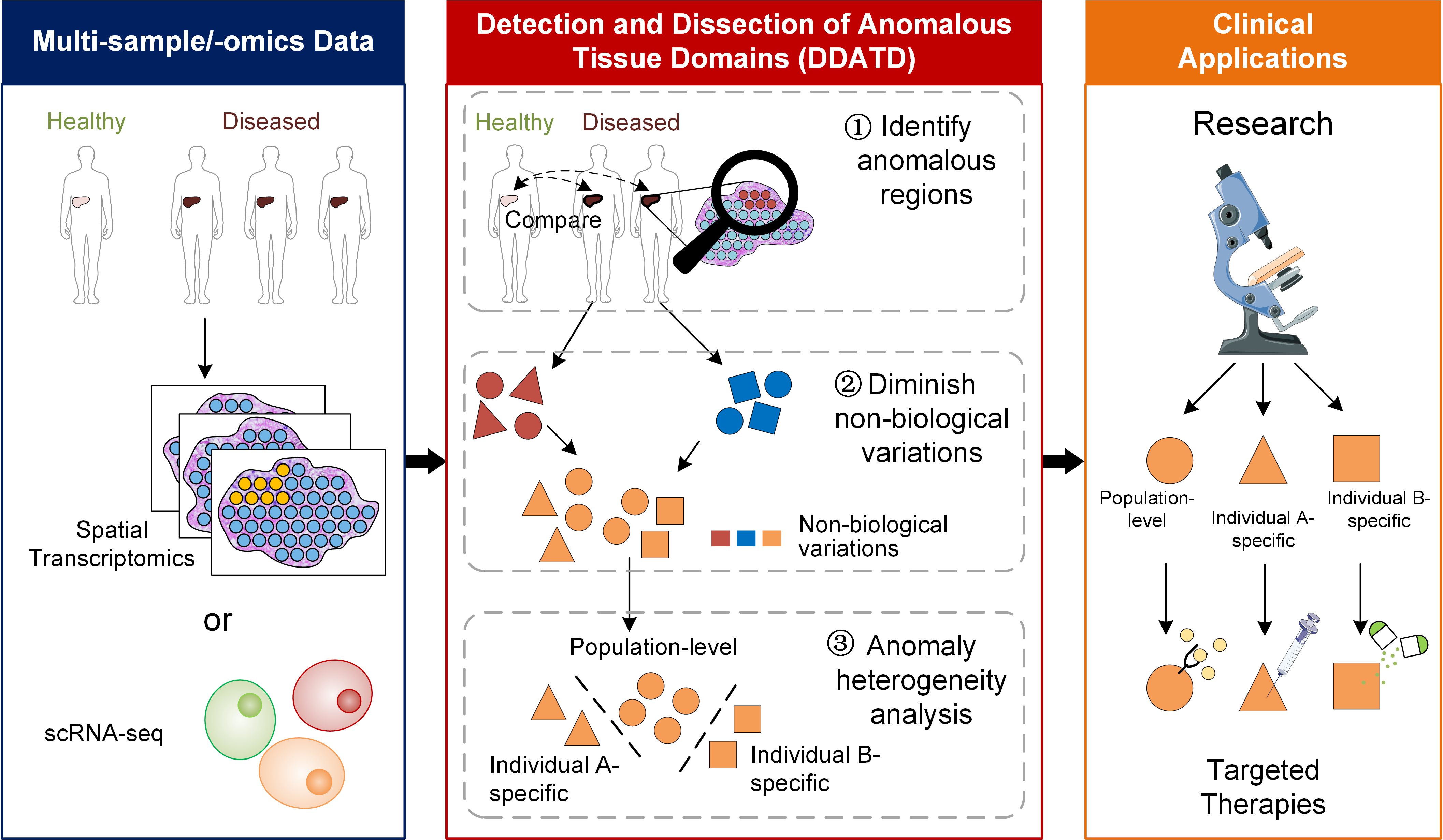
Domain Adaptive and Fine-grained Anomaly Detection for Single-cell Sequencing Data and Beyond
ACSleuth is a GAN-based generative model for domain adaptive and fine-grained anomaly detection in the single-cell/tabular data. 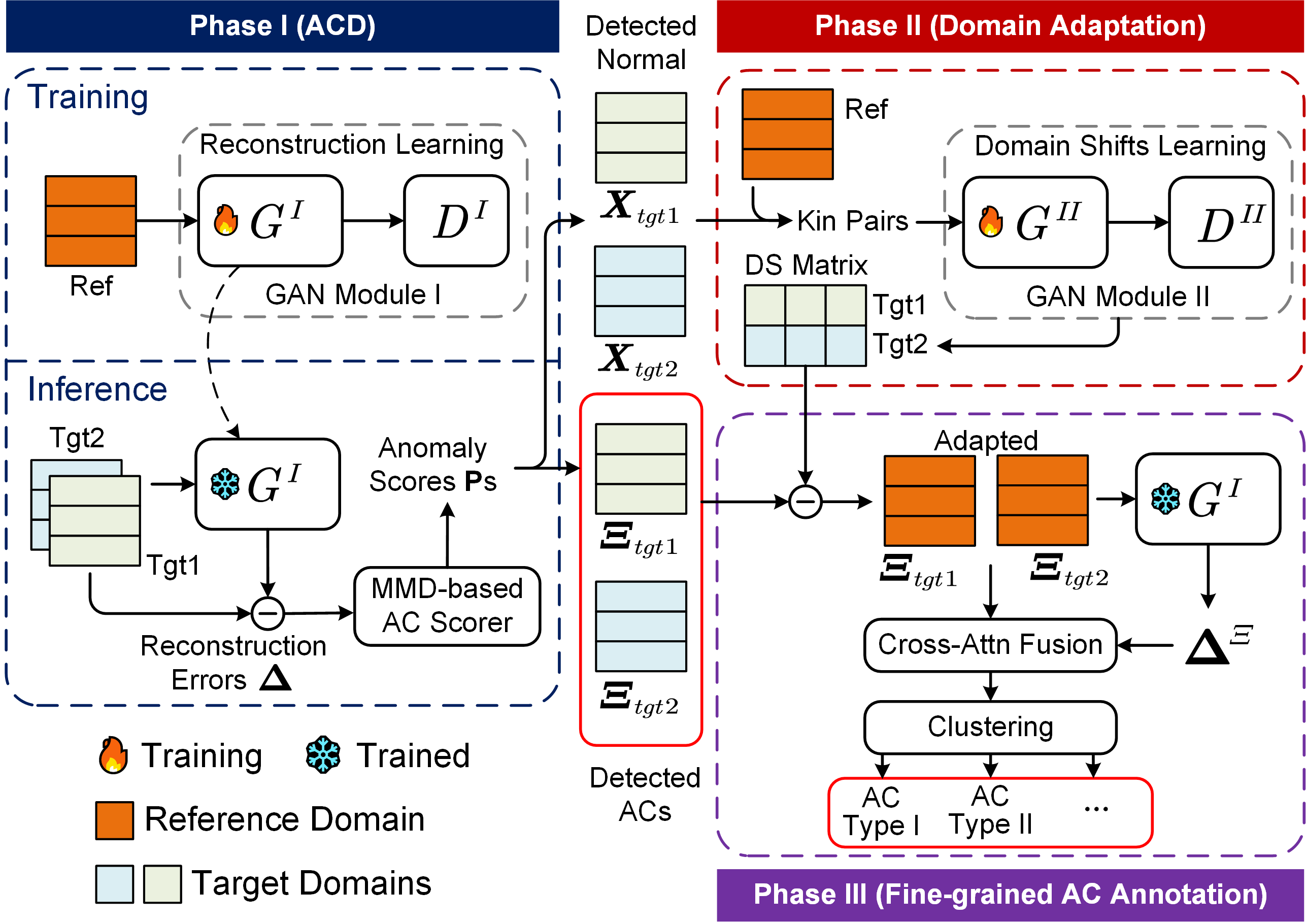
publications
talks
Oral Academic Seminar about Our Work ACSleuth
Published:
We discussed how to detect anomalous samples in single-cell omics data with generative models and introduced our team’s work, ACSleuth.
Oral Academic Presentation about Our Work ACSleuth
Published:
We presented our team’s work, ACSleuth, discussing in detail its implementation specifics, applicable tasks, and the limitations of the algorithm.
Xiaoxiang Neurology Rare Disease Academic Salon
Published:
We introduced multimodal machine learning technologies for multi-omics data, and discussed how to discover rare diseases with deep learning methods.
Oral Academic Seminar about Our Work STANDS and ACSleuth
Published:
We discussed how to detect anomalous samples in multi-omics data with generative models and introduced our team’s work, ACSleuth and STANDS.
Oral Academic Presentation about Our Work ACSleuth
Published:
We formally introduced our work, Domain Adaptive and Fine-grained Anomaly Detection for Single-cell Sequencing Data and Beyond.
